Nucleotide sugar production in plants
|
||
AGP degrading enzymes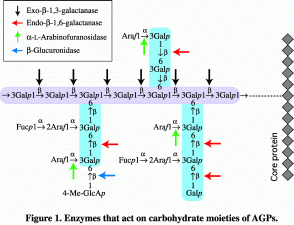
Arabinogalactan-proteins (AGPs) are a family of complex
proteoglycans found in all tissues of higher plants and localized in
cell walls, plasma membranes, and the extracellular matrix. Although
the functions of AGPs have not been clearly identified, several lines
of evidence indicate that they are involved in many physiological
events such as cell division, cell expansion, and cell death. AGPs are
characterized by large amounts of carbohydrate components rich in
galactose and L-arabinose, and protein components (core
proteins: generally less than 10% of total weight) rich in
hydroxyproline, serine, threonine, alanine, and glycine. The reducing
galactosyl residues in the carbohydrate chains are attached through O-glycosyl
linkages to Hyp and/or Ser/Thr residues. Since a large number of
putative protein cores exist, it is difficult to separate particular
AGP molecules from other AGP species in plant tissues, which make it
difficult to elucidate the precise characteristics of individual AGPs.
The carbohydrate moieties of AGPs have a common structure consisting of
ƒÀ-1,3-galactosyl backbones to which side chains of ƒÀ-1,6-galactosyl
residues are attached through O-6. L-Arabinose
and lesser amounts of other auxiliary sugars such as glucuronic acid, 4-O-methyl-glucuronic
acid, L-rhamnose, and L-fucose are
attached to the side chains, usually at nonreducing terminals. It is
important to study carbohydrate degrading enzymes of AGPs because
hydrolytic enzymes specific to particular sugar residues and type of
glycosidic linkage provide useful tools for structural analysis of the
sugar moieties of AGPs (Figure 1). To
date we have characterized several hydrolytic enzymes such as
exo-ƒÀ-1,3-galactanase (EC 3.2.1.145), endo-ƒÀ-1,6-galactanase (EC
3.2.1.164), endo-ƒÀ-1,3-galactanase (EC
3.2.1.181), ƒÀ-galactosidase (EC 3.2.1.23), ƒ¿-L-arabinofuranosidase
(EC 3.2.1.55), and ƒÀ-glucuronidase (EC 3.2.1.31). ReferencesImaizumi et al. (2017) J.
Exp. Bot. 68: 4651-4661 (abstract)
Yoshimi et al. (2017) Carbohydr. Res. 453-454: 26-35 (abstract) Kitazawa et al. (2013) Plant Physiol. 161: 1117-1126 (abstract) Kotake et al. (2011) J. Biol. Chem. 286: 27848-27854 (abstract) Konishi et al. (2008) Carbohydr. Res. 343: 1191-1201 (abstract) Ichinose et al. (2008) Appl. Environ. Microbiol. 74: 2379-2383 (abstract) Kotake et al. (2006) J. Exp. Bot. 57: 2353-2362 (abstract) Kotake et
al. (2005) Plant Physiol. 138: 1563-1576 (abstract)
Ichinose et al. (2005) J. Biol. Chem. 280: 25820-25829 (abstract) Kotake et
al. (2004) Biochem. J. 377:
749-755 (abstract) |
||